Find gaps and overlaps in global topological boundaries
Gaps and overlaps are caused when:
there is an area of the globe not covered by a topological boundary or network, or
two (or more) topological boundary polygons overlap in some area of the globe.
This can also happen if two topological line sections are identical when ideally there should only be one of them (and it should be shared by two neighbouring topological boundaries).
Sample code
import pygplates
# Create a topological model from the topological plate polygon features (can also include deforming networks)
# and rotation file(s).
topological_model = pygplates.TopologicalModel('topologies.gpml', 'rotations.rot')
# Our geological times will be from 0Ma to 'num_time_steps' Ma (inclusive) in 1 My intervals.
num_time_steps = 140
# 'time' = 0, 1, 2, ... , 140
for time in range(num_time_steps + 1):
# Get a snapshot of our resolved topologies at the current 'time'.
topological_snapshot = topological_model.topological_snapshot(time)
# Extract the boundary sections between our resolved topological plate polygons (and deforming networks) from the current snapshot.
shared_boundary_sections = topological_snapshot.get_resolved_topological_sections()
# We'll create a feature for any anomalous sub-segment we find.
anomalous_features = []
# Iterate over the shared boundary sections.
for shared_boundary_section in shared_boundary_sections:
# Iterate over the sub-segments that actually contribute to a topology boundary.
for shared_sub_segment in shared_boundary_section.get_shared_sub_segments():
# If the resolved topologies have global coverage with no gaps/overlaps then
# each sub-segment should be shared by exactly two resolved boundaries.
if len(shared_sub_segment.get_sharing_resolved_topologies()) != 2:
# We keep track of any anomalous sub-segment features.
anomalous_features.append(shared_sub_segment.get_resolved_feature())
# If there are any anomalous features for the current 'time' then write them to a file
# so we can load them into GPlates and see where the errors are located.
if anomalous_features:
# Put the anomalous features in a feature collection so we can write them to a file.
anomalous_feature_collection = pygplates.FeatureCollection(anomalous_features)
# Create a filename (for anomalous features) with the current 'time' in it.
anomalous_features_filename = 'anomalous_sub_segments_at_{0}Ma.gpml'.format(time)
# Write the anomalous sub-segments to a new file.
anomalous_feature_collection.write(anomalous_features_filename)
Details
topological model from topological features and rotation files.pygplates.RotationModel) can be specified here,
however we’re only specifying a single rotation file.topological_model = pygplates.TopologicalModel('topologies.gpml', 'rotations.rot')
Note
We create our pygplates.TopologicalModel outside the time loop since that does not require time.
time
using pygplates.TopologicalModel.topological_snapshot().topological_snapshot = topological_model.topological_snapshot(time)
pygplates.ResolvedTopologicalBoundary (used for dynamic plate polygons) and
pygplates.ResolvedTopologicalNetwork (used for deforming regions) are listed in the boundary sections.shared_boundary_sections = topological_snapshot.get_resolved_topological_sections()
These boundary sections are actually what
we’re interested in because their sub-segments have a list of topologies on them.
pygplates.ResolvedTopologicalSection.get_shared_sub_segments().for shared_sub_segment in shared_boundary_section.get_shared_sub_segments():
sub-segment
is obtained using pygplates.ResolvedTopologicalSharedSubSegment.get_sharing_resolved_topologies.if len(shared_sub_segment.get_sharing_resolved_topologies()) != 2:
If a sub-segment is not shared by exactly two resolved boundaries then we record its feature.
anomalous_sub_segment_features.append(shared_sub_segment.get_resolved_feature())
Finally we write the anomalous features to a file.
anomalous_features_filename = 'anomalous_sub_segments_at_{0}Ma.gpml'.format(time)
anomalous_feature_collection.write(anomalous_features_filename)
Visualising gaps and overlaps in GPlates
The resulting output files such as anomalous_sub_segments_at_10Ma.gpml can be loaded into
GPlates to see where the topological errors are located on the globe.
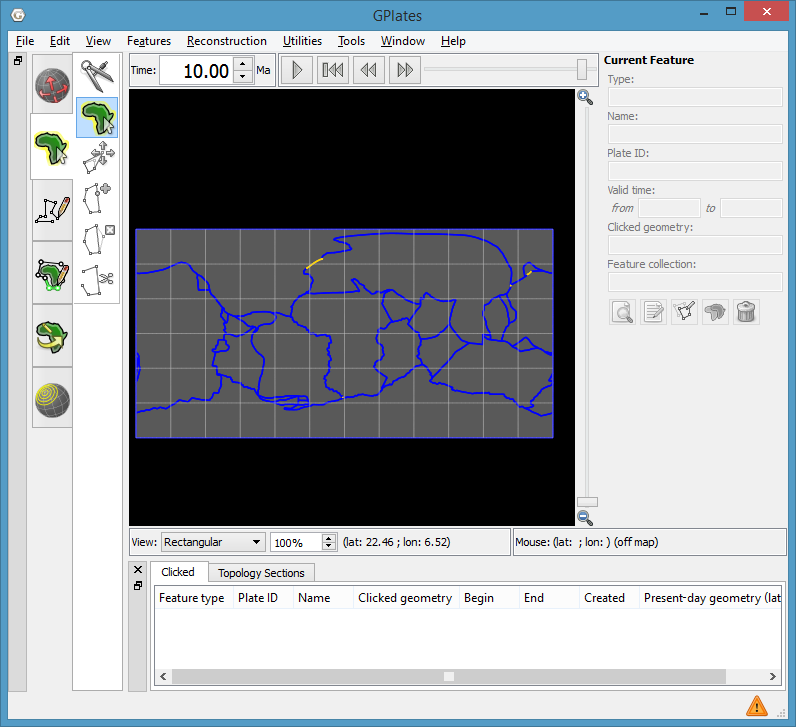
GPlates screenshot showing anomalous sub-segments (yellow) and dynamic plate polygons (blue) at 10Ma.
The following two screenshots show a zoomed-in view of a gap and an overlap.
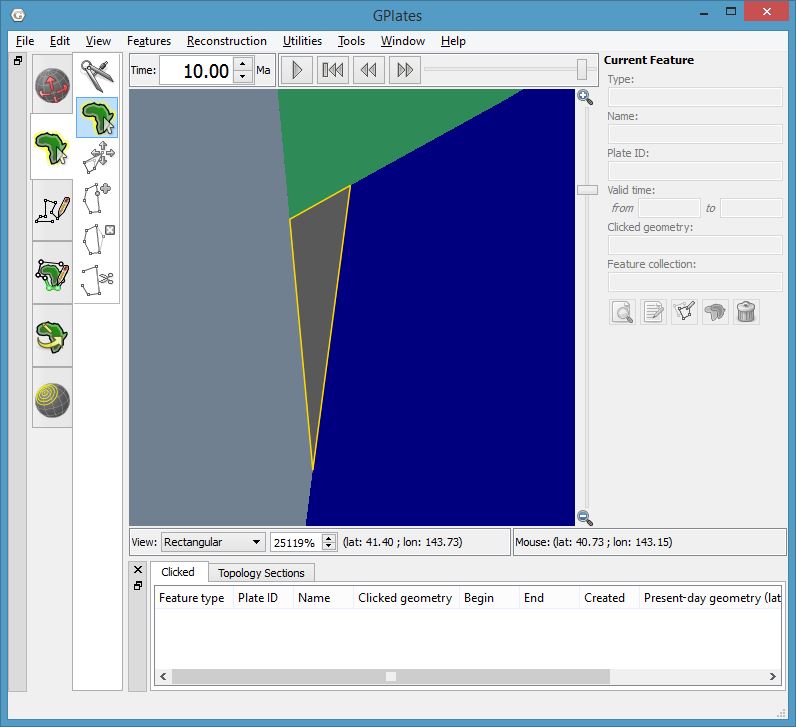
GPlates screenshot showing zoomed-in view of a gap in dynamic polygon coverage (outlined in yellow) at 10Ma.
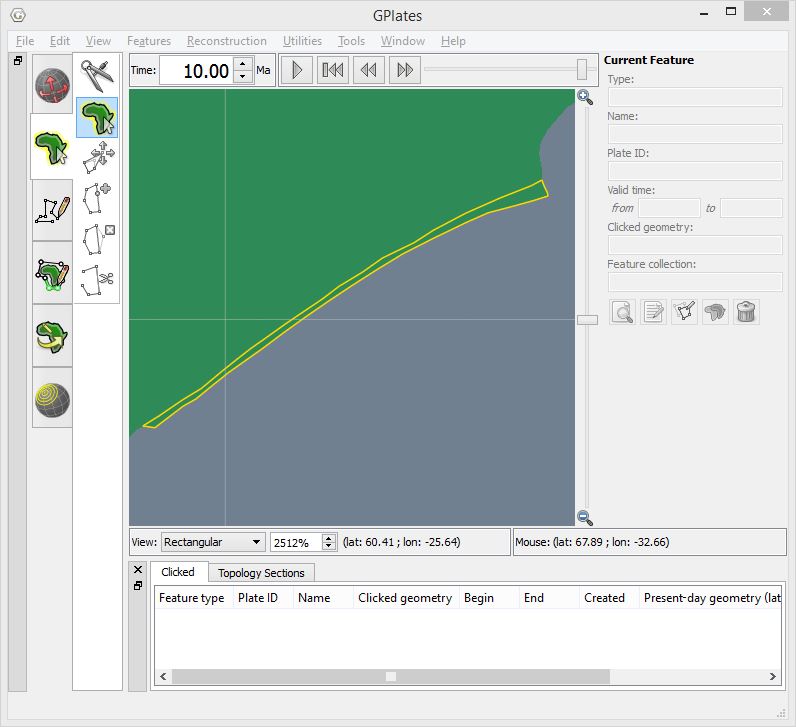
GPlates screenshot showing zoomed-in view of an overlap in dynamic polygon coverage (outlined in yellow) at 10Ma.